Selected Recent Publications
Quantifying metabolites using structure-switching aptamers coupled to DNA sequencing. Tan JH, Fraser AG. Nat Biotechnol. 2025 Feb 4; online ahead of print.
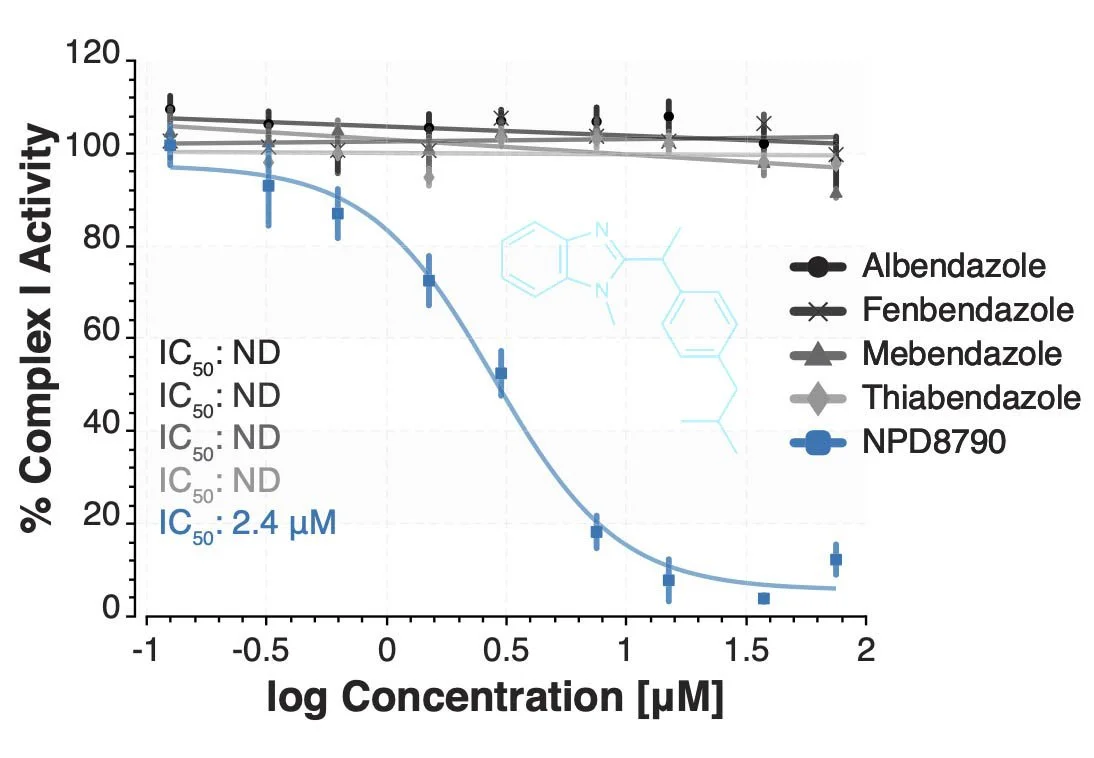
Identification of a family of species-selective complex I inhibitors as potential anthelmintics. Davie T, Serrat X, Imhof L, Snider J, Štagljar I, Keiser J, Hirano H, Watanabe N, Osada H, Fraser AG. Nat Commun. 2024 May 8; 15(1):3367.
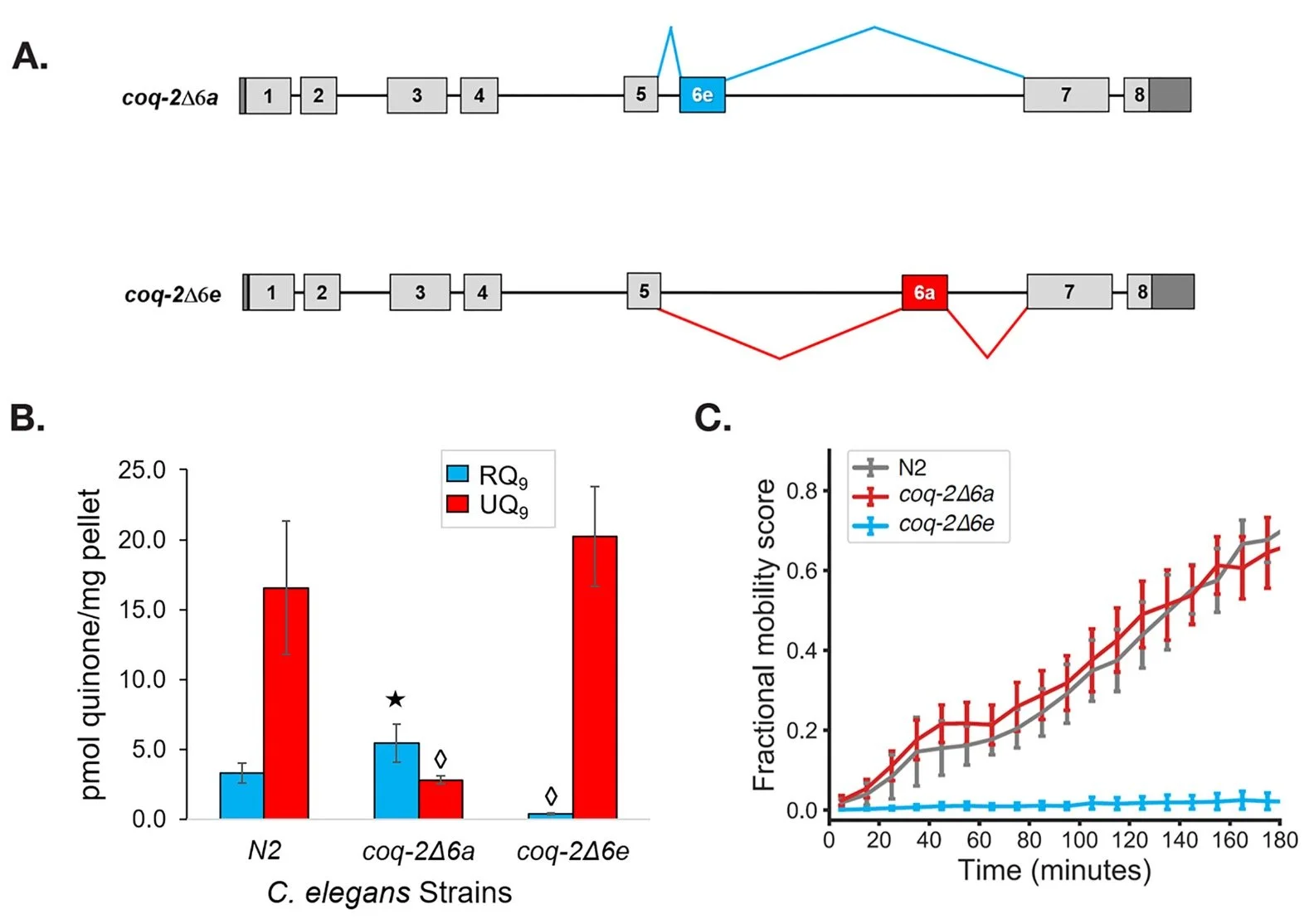
Alternative splicing of coq-2 controls the levels of rhodoquinone in animals. Tan JH, Lautens M, Romanelli-Cedrez L, Wang J, Schertzberg MR, Reinl SR, Davis RE, Shepherd JN, Fraser AG, Salinas G. eLife. 2020 Aug 3; 9:e56376.
Rhodoquinone biosynthesis in C. elegans requires precursors generated by the kynurenine pathway. Del Borrello S, Lautens M, Dolan K, Tan JH, Davie T, Schertzberg MR, Spensley MA, Caudy AA, Fraser AG. Elife. 2019 Jun 24;8. pii: e48165.
Acute Effects of Drugs on Caenorhabditis elegans Movement Reveal Complex Responses and Plasticity.
Spensley M, Del Borrello S, Pajkic D, Fraser AG.
G3 (Bethesda). 2018 Aug 30;8(9):2941-2952.
The combinatorial control of alternative splicing in C. elegans.
Tan JH, Fraser AG.
PLoS Genet. 2017 Nov 9;13(11):e1007033.
Taxonomically Restricted Genes with Essential Functions Frequently Play Roles in Chromosome Segregation in Caenorhabditis elegans and Saccharomyces cerevisiae.
Verster AJ, Styles EB, Mateo A, Derry WB, Andrews BJ, Fraser AG.
G3 (Bethesda). 2017 Oct 5;7(10):3337-3347.
Natural Variation in Gene Expression Modulates the Severity of Mutant Phenotypes.
Vu V, Verster AJ, Schertzberg M, Chuluunbaatar T, Spensley M, Pajkic D, Hart GT, Moffat J, Fraser AG.
Cell. 2015 Jul 16;162(2):391-402.
Comparative RNAi screens in C. elegans and C. briggsae reveal the impact of developmental system drift on gene function.
Verster AJ, Ramani AK, McKay SJ, Fraser AG.
PLoS Genet. 2014 Feb 6;10(2):e1004077.
The majority of animal genes are required for wild-type fitness.
Ramani AK, Chuluunbaatar T, Verster AJ, Na H, Vu V, Pelte N, Wannissorn N, Jiao A, Fraser AG.
Cell. 2012 Feb 17;148(4):792-802.