RESEARCH OVERVIEW
Metabolites are critical biomarkers — the levels of glucose, amino acids, hormones, and vitamins give us a snapshot of our health and errors in metabolite levels cause diseases like diabetes and phenylketonuria. However, it is hard to quantify metabolites because they are biochemically highly diverse and cannot be amplified. How can we quantify this huge range of molecules in samples ranging from tissues to plasma to single cells in a rapid, efficient manner?
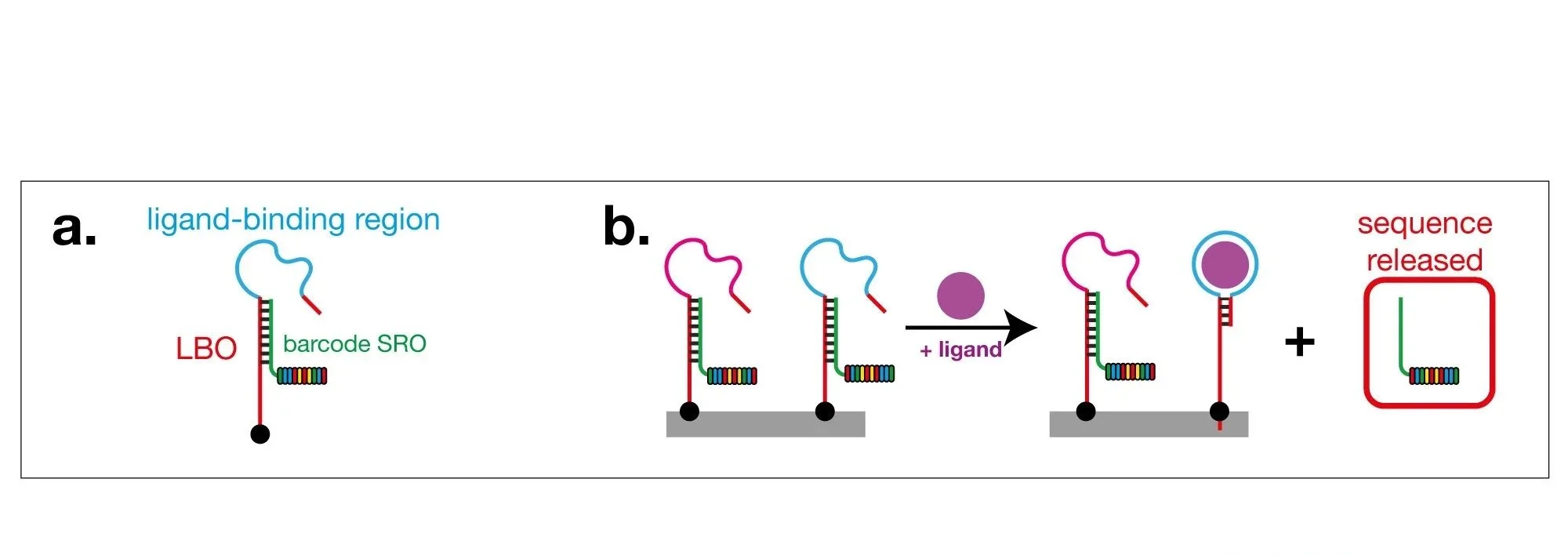
Our current research focuses on harnessing the incredible power of DNA sequencing to revolutionise the detection and measurement of metabolites and drugs. We recently developed a method that uses DNA sequencing to read metabolite and drug concentrations. This method, that we call smol-seq (for Small MOLecule sequencing), uses structure-switching aptamers (SSAs) as sensors to detect individual targets. Each SSA detects a single target and is coupled to a unique DNA barcode — when the target binds, this causes the release of the barcode e.g. a glucose SSA ‘sees’ glucose and releases a ‘glucose’ barcode and an SSA that ‘sees’ phenylalanine releases a different ‘phenylalanine’ barcode. Simply by reading and quantifying the released barcoded SROs, it is thus possible to quantify the metabolites in a complex mixture.
We showed these SSAs are highly specific, distinguishing between closely related targets, that the level of barcode release gives a quantitative readout of target concentration, and that many of these SSAs can be read in parallel since each has a different barcode. smol-seq has the potential to completely change metabolomics and to allow researchers to integrate metabolite measurements into ‘multiomics’ platforms — it is ‘next-gen metabolomics’. Our goal over the next 5 years is to establish smol-seq as a fully plug-and-play platform for metabolomics by DNA sequencing.
To get a sense of the foundation of this work in greater detail see our paper here:
Tan JH and Fraser AG. Quantifying metabolites using structure-switching aptamers coupled to DNA sequencing. Nature Biotechnology. 2025
SPECIFIC RESEARCH GOALS
1. Develop sensors for a set of ~200 key metabolites. This will allow us to simultaneously measure core metabolites like amino acids, sugars, steroid hormones, nucleotides, and vitamins using DNA sequencing and establish smol-seq as a new metabolomics platform.
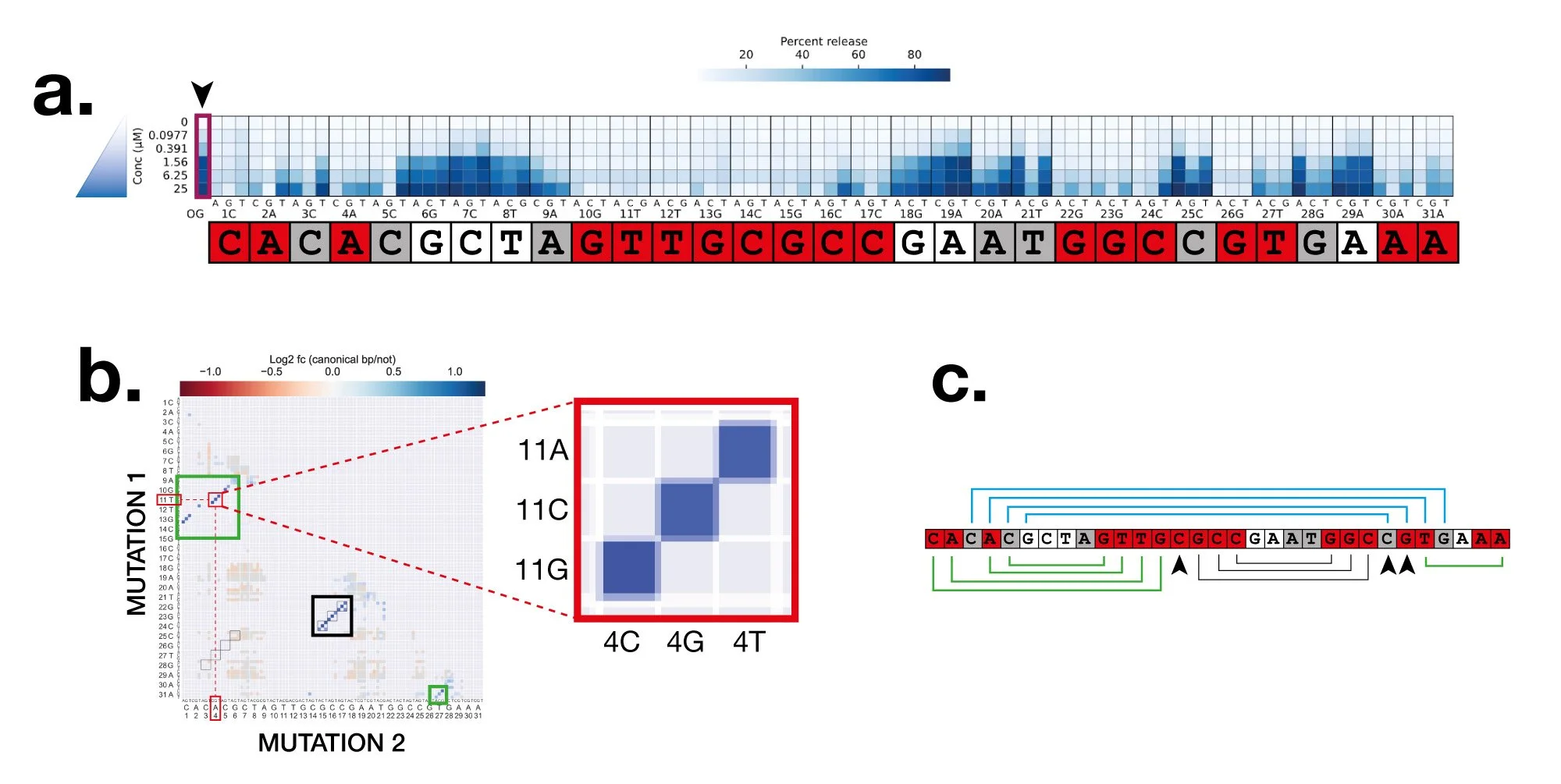
2. Understand how SSAs work. How does each SSA specifically recognise its target and what is the exact conformational change it undergoes when the target binds? Can we alter the specificity and sensitivity in knowledge-directed ways? We tackle these using a variety of methods including deep mutational scanning and massively parallel functional assays — these give rich insights into the basic mechanisms underlying SSA-target interactions. This is a pure biology goal — SSAs and aptamers in general are really cool molecules and for many of them, we really don’t understand how they work. Yet.
Develop computational models to predict SSAs for novel targets. The combination of massive data and computational methods has unlocked incredibly complex biological problems like protein structure prediction. We aim to build massive datasets measuring how individual SSAs recognise a wide range of targets and use these to train models to predict SSAs de novo for novel targets. This would be transformative — if we can predict a sensor for any target of interest it would open up incredible new science.